Numpy Tricks and A Strong Baseline for Vector Index
I like simple things and not dependencies. When it comes to architecting Jina, I keep its core dependencies as simple as numpy, pyzmq, protobuf, and grpcio while still delivering full-stack functionalities. This allows Jina developers & users to bootstrap an end-to-end neural search system without extra packages quickly.
On the vector indexing and querying part, Jina has implemented a baseline vector indexer called NumpyIndexer, a vector indexer that is purely based on numpy. The implementation pretty straightforward: it writes vectors directly to the disk and queries nearest neighbors via dot product. It is simple, requires no extra dependencies, and the performance was reasonable on small data. As the default vector indexer, we have been using it since day one when showcasing quick demos, toy examples, and tutorials.
Recently, this community issue has raised my attention. I realize there is a space of improvement, even for this baseline indexer. In the end, I manage to improve the index and query speed by 1.6x and 2.8x while keeping the memory footprint constant (i.e., invariant to the size of the index data). This blog post summarizes the tricks I used.
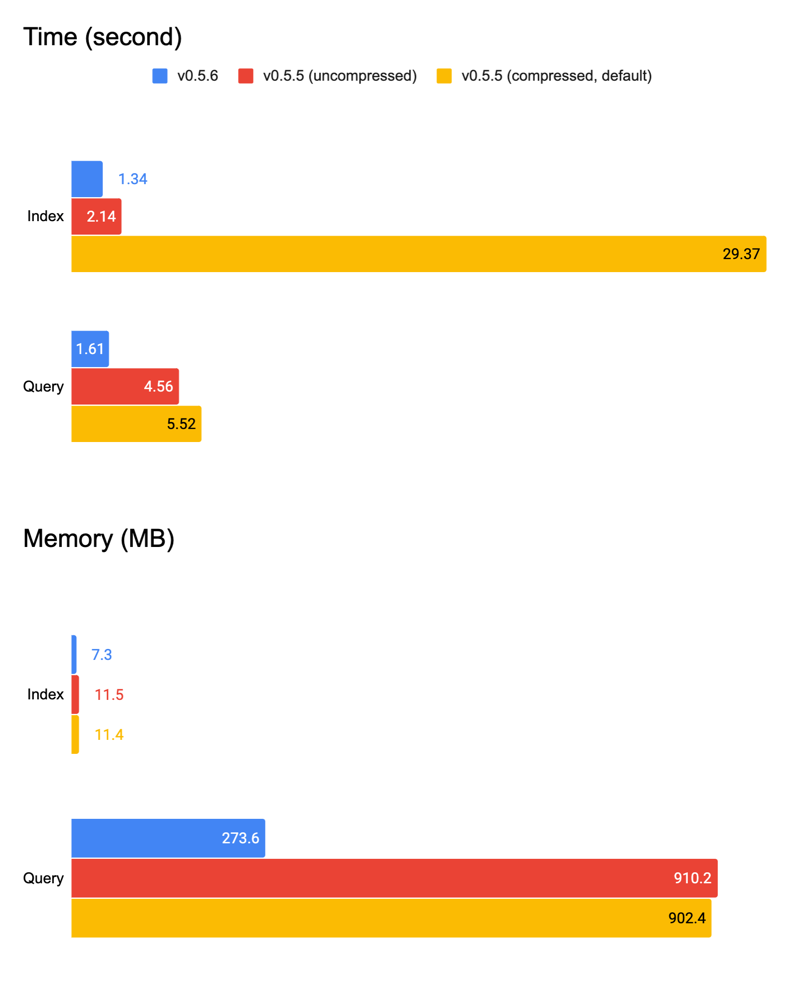
Table of Contents
- The Scalability Problem
- numpy.memmap Instead of numpy.frombuffer
- Batching with Care
- Memory-efficient Euclidean and Cosine
- Removing gzip compression
- Summary
The Scalability Problem
As the issue points out, NumpyIndexer faces a severe scalability problem: in the query time, it tries to load all data into memory, making it (and all inheritances) unusable on big data. If the user has 8GB physical memory on its laptop, then the biggest index size that NumpyIndexer can support is around 8GB, which is 7 digits embeddings at most. Having this in mind, my solution is built on memmap naturally.
numpy.memmap Instead of numpy.frombuffer
Replacing the full-read to numpy.memmap is trivial. Thanks to the ndarray format Jina followed in the index time; the only necessary work is changing how raw_ndarray is built at the query time.
| Before | After | ||||
|---|---|---|---|---|---|
|
|
This reduces loading time basically to zero, as ndarray is now just a pointer to the virtual memory, it can be built in no time. The data behind this pointer is loaded on demand. Simple ops such as ndarray[1], ndarray.shape won’t trigger the full-read.
Batching with Care
To query nearest neighbors NumpyIndexer computes dot products between query vectors and all indexed vectors row-wise. That means it anyway has to scan over the full data.If we stop here and don’t optimize any further, then the memory consumption is back to the high watermark after the first scan. The correct way is adding “batching” to all computations, e.g. dot products, cosine distance. Jina has provided a powerful @batching decorator from day one, and here is how to use it:
| Before | After | ||||
|---|---|---|---|---|---|
|
|
Lifecycle of memmap
This decorator will slice raw_B into batches and compute _euclidean one by one. All partial results are then merged before returning. However, it didn’t work as expected. Below is the memory footprint when running 100 x 10,000 query against 10,000 x 10,000 indexed vectors:
1 | Line # Mem usage Increment Line Contents |
One can observe that all data is loaded after the first query. Though each time batch_euclidean only works with partial data, the program eventually loads all data into the cache buffer. This buffer (as part of the virtual memory) is controlled on the OS level. There are arguments that claim this buffer will be released by OS automatically when the memory is intense. I decided to manually release this buffer by restricting the lifecycle of memmap inside the batching for-loop.
1 | data = np.memmap(self.index_abspath, dtype=self.dtype, mode='r', shape=(self.size, self.num_dim)) |
Zero-copy slicing
When generating slice, the previous Jina uses numpy.take. One gotcha of numpy.take is it copies the data instead of slicing in-place, costing extra memory. One can use Python built-in slice function to implement zero-copy version of numpy.take as follows:
numpy.take | built-in slice (zero-copy) | ||||
|---|---|---|---|---|---|
|
|
Memory-efficient Euclidean and Cosine
When computing Euclidean and Cosine distance row-wise on two matrices, one can simply use the broadcast feature of ndarray:
1 | numpy.sqrt(((A[:,:,None]-B[:,:,None].T)**2).sum(1)) |
However, [:,:,None] - [:,:,None].T creates a (num_query, num_dim, num_data) matrix in memory and this method can quickly become infeasible on big data. The space complexity is $O(N^3)$. In Jina, we use a more memory-efficient way to compute Euclidean distance:
1 | def _ext_A(A): |
The idea is simply doing $(a-b)^2 = a^2 -2ab +b^2$ explicitly: first prefilling the values of $a^2$, $b^2$, $a$, $-2b$, and then computing $-a \times 2b$ part. This reduces the memory to (num_query*3, num_dim) + (num_data*3, num_dim) and the overall space complexity to $O(N^2)$.
Notice that when computing _euclidean with the @batching decorator, A is the query matrix and remains constant across all batching iterations. Hence, A_ext can be precomputed before the batching loop to avoid unnecessary computation.
Finally, the Cosine distance is nothing more but the “normed” Euclidean distance. It can be simply built on top of _euclidean function by feeding _norm(A) and _norm(B) to it:
1 | def _norm(A): |
Removing gzip compression
Jina uses gzip to store the vector data. This has been an old feature since the day one. However, this feature does not play well with memmap because the decompression will have to load all data into memory and this is against the idea of on-demand computing.
While removing gzip I found it didn’t add many benefits in the first place. The table below summarizes with/without gzip compression on v0.5.5 (this version does not include any improvement mentioned abov). One can see that the index time increases 15x, whereas results in only 40MB space-saving.
v0.5.5 Mode | Time (second) | On-disk Space (MB) |
|---|---|---|
| Index | 2.13 | 800 |
| Index (compressed) | 29.36 | 759 |
| Query | 4.56 | |
| Query (compressed) | 5.52 |
The script for benchmarking can be found here.
Summary
Putting everything together, now we get a pretty good baseline vector indexer that uses constant memory. We have rolled out this improved NumpyIndexer in v0.5.6 patch and all examples, tutorials, and jina hello-world can immediately enjoy this improvement.
| Version | Time | Memory | ||
|---|---|---|---|---|
| Index | Query | Index | Query | |
v0.5.6 | 1.34s | 1.61s | 7.3MB | 273.6MB |
v0.5.5 | 2.14s | 4.56s | 11.5MB | 910.2MB |
v0.5.5 (compressed) | 29.36s | 5.52s | 11.4MB | 902.4MB |
To me the use of memmap is the most inspiring part. It makes me think a lot. This function and memoryview() can be used in Jina more extensively to improve its scalability further.